As the spread of the novel coronavirus continues, interest in the development of treatments is also increasing rapidly. According to the conventional methodologies of drug discovery, a new drug is developed through an exhaustive process in which new candidate substances are screened using the knowledge of experts and tested through clinical trials.
However, these traditional methods of drug discovery are very costly in terms of time and money. Early studies for new drug discovery can take a relatively long period of 2-3 years, and to go through clinical trials to make final placement in the market, it could easily take up to 10 years. Recent estimations calculate that the entire process costs between $3 to 4 billion on average, and above all, the success rate is extremely low. Only one or two of every 10,000 tested molecules will successfully become a marketable medicine.
In contrast, the outbreak cycle of new infectious diseases around the world is getting shorter. To overcome this, the cycle of developing new medicines must be accelerated and several methods are put into operation. Among them, Artificial Intelligence (AI) technologies are at the center of the most recent trends in developing marketable medicines.
Many companies have been applying AI to speed up the overall process of drug development. Resources have been piling up in the pharma field and AI-based drug-discovery startups have seen a rise in funding since 2018. In fact, every one of the major pharmaceutical companies has announced a partnership with at least one startup to facilitate an expedited drug discovery process. To get a glimpse of these companies' technological prowess in drug discovery, let’s take a look at their patents.
From the patents of drug developers utilizing AI, you can find that the AI algorithms used for drug development are not completely new ones, specially designed to serve the purpose of drug development. Rather, it is about how to process biological data sets to feed already proven algorithms so that the computer can provide solutions to the pharmaceutical industry.
Many people assume that filing AI-related patents is only possible when creating a new model or algorithm. However, there exists an independent category for patents related to AI data processing that can be registered if there is novelty and inventiveness in the methods.
To apply the algorithm to a specific field of research, it is necessary to somehow process the data in the proper way to feed AI-based models to provide results for those data sets in the specific field. As you can imagine, this is valid for any type of data, and so patent application on data processing is open for every industry.
Such trend can be particularly noticed in the pharmaceutical industry, since each performance of AI-based drug discovery itself showcases its own distinctiveness according to how the data sets are processed, as they involve technical data sets.
Throughout this article, we will take a look at patents of AI-technologies applied to the pharma industry from four drug discovery startups. First, we will take a look at U.S.-based Atomwise followed by three Korean companies; Standigm, MediRita and Syntekabio.
1. Atomwise
Atomwise is a San Francisco-based startup that developed AtomNet, a system that uses AI and machine learning algorithms for structure-based drug design and discovery. The company became the center of attention when it signed a $550 million dollar contract with the pharma giant, Eli Lily in 2019.
Now let’s take a look at their “hot and trending” patent “Systems and methods for applying a convolutional network to spatial data” (US 10482355).
To briefly explain, this patent refers to three-dimensional voxel maps fed into a convolutional neural network (CNN) to estimate the binding affinity between the test object (e.g. a chemical compound) and the target object (e.g. protein, polymer).
To be more specific, the features obtained by inputting a three-dimensional voxel map to the convolutional layer are input to the fully connected layer to obtain a composite score that indicates the binding affinity score of the test object and the target object. The overall idea of this patent is to classify test objects into classes according to the binding affinity scores, and finally determine which class the test object belongs to.
Those accustomed to feeding two-dimensional images into CNNs are probably wondering how to enter three-dimensional spatial data into CNNs and what form of data a voxel map is. Hence, I will explain it below.
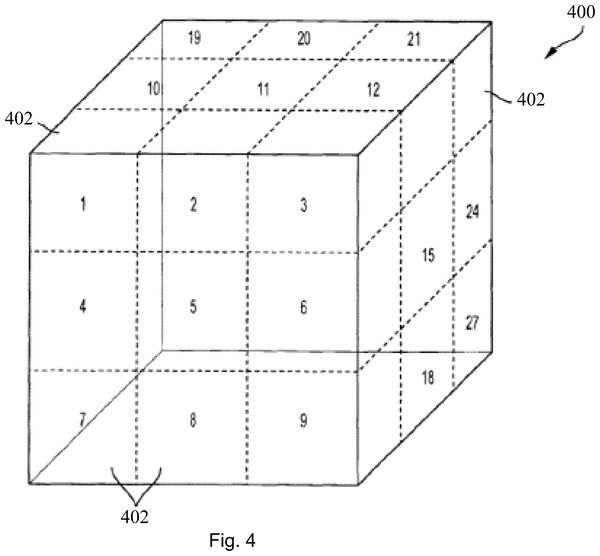
In a nutshell, in this patent, the voxel maps are converted into suitable forms and sequentially fed into the convolutional neural network. Each voxel is a volume element that represents the target object on a three-dimensional grid basis. Also, each voxel map consists of the test object (e.g. chemical compounds) and the active site of the target object (e.g. protein, polymers) in respective poses among the plurality of different poses. To explain in detail the vortex map, the active portions of the polymer are aligned to be located at the center of the voxel map, and the voxel map represents a variety of compositions with different poses with the active portion aligned in the center. Thereby, creating a plurality of poses for the target object.
Now, how does the voxel map represent the information about the target object or the test object? Each voxel in the voxel map expresses information about the target or test object by expressing each voxel in a different color depending on which atom is located. In another embodiment, each voxel may be represented by an atomic number located on the voxel.
Schematic view of the geometric representation of input features in the form of a 3D grid of voxels.
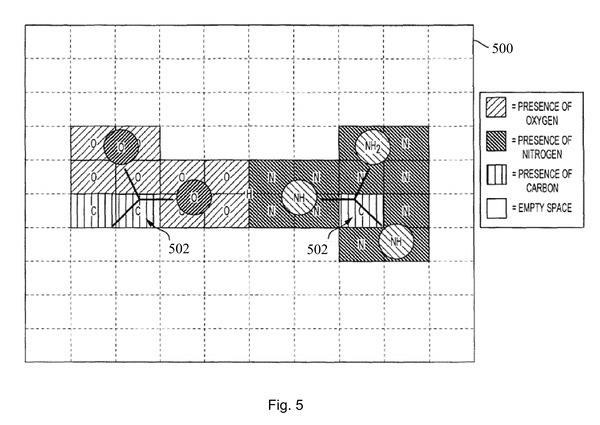
View of two objects encoded onto a two-dimensional grid of voxels, different colors are expressed depending on the atom.
Atomwise’s patent uses the CNN without major modifications to predict the binding preference between a test compound and the target protein. Therefore, it may seem that there is no technological improvement.
However, the technological feature of this patent lies in that the input data is composed of a three-dimensional voxel map to use a CNN to predict the binding affinity of the test compound to the target protein.
Based on this, we were able to look at the concerns and methods on how to organize the data input to a general AI algorithm through the Atomwise patent.
Now, let’s learn about how Korean companies enter data into artificial intelligence algorithms.
2. Standigm
First, let’s take a look at Standigm. Standigm is a startup based in Seoul, South Korea, founded in 2012 that designs new drugs utilizing artificial intelligence.
Standigm attracted attention in 2019 when the Korean conglomerate, SK Holdings announced an investment of KRW 10 billion (US$8.6 million) for AI-based drug development.
The patent from Standigm, examined in this article, is “Method for predicting therapeutic efficacy of the combined drug by machine learning ensemble model” (US20180060483A1).
Combined drugs are developed by combining preexisting drugs. Such combined drugs can produce synergistic effects that could lead to reducing the risk of side effects because they could show greater effect with smaller concentrations.
Standigm’s patent presents a method that uses machine learning algorithms to evaluate the therapeutic efficiency of combined drugs.
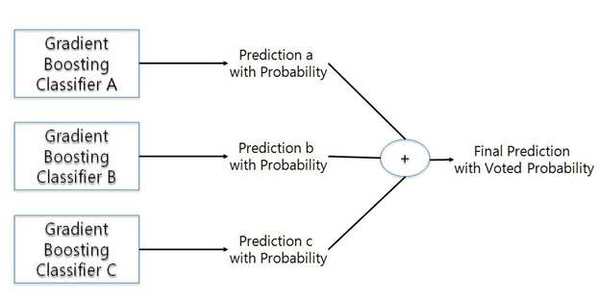
Standigm’s patent predicts the therapeutic efficacy of a combined drug by predicting the combined therapeutic efficacy of drugs using an ensemble of a plurality of gradient boosting classifier models as sequential ensemble learning. The gradient boosting model is a classifier model made with an ensemble of several weak prediction models, typically decision trees, to improve the prediction performance. In this patent, the model calculates the probability value determining whether or not the drug combinations have a synergic effect.
For an easier explanation of the process to evaluate the efficiency of combination drugs, we will use the following example. For a specific combined drug, gradient boosting model A outputs a confidence value of 0.96 (96% probability of synergy effect of combination drugs) and gradient boosting model B calculates a confidence value of 0.84 (probability of synergy of combined drugs, 84%). In this case, the output values in the gradient boosting model A and B are summed up to evaluate the effect of the combined drug.
Multiple gradient boosting model.
As such, this patent from Standigm predicts the effect of combination drugs by simply using a gradient boosting model.
However, the essence of this patent can be seen in the input data fed into the model, which is described below. Thus, let’s take a look at how Standigm designed the input data.
In this patent, the input data is expressed by matrices. Among the input data, the data at the gene level is converted into the form of map-based matrices with genes as rows and samples as columns, as shown in the figure below. For example, a mutation matrix at a gene level contains data related to mutations at the ‘gene level’. The mutation matrix is displayed as ‘1’ when the mutation-related data provided at the gene level includes mutations and as predicted value ‘0' when it includes no mutations.
Mutation matrix at a gene level.
That way, the first step of the method provides cell-related data, drug-related data and drug/cell correlation-related data as the ‘gene-level data’. Then, in the second step, it provides ‘pathway-level data’ which is deduced from the gene-level data. Since pathway-level data is extracted from gene-level data, it can be viewed as pre-processed data and represented as a matrix, with paths as rows and samples as columns.
Therefore, in summary, it can be said that in this patent, the pre-processed ‘pathway map module’ is fed into a computer learning algorithm (gradient boosting model) to predict the effects of combination drugs.
As mentioned earlier, the patent of Standigm is deemed to be patentable in that it is the process of inputting complex cells and drug data into an AI model.
3. MediRita
MediRita, founded in 2018, is another Korean startup that develops new drug development technologies with artificial intelligence.
MediRita applied for a patent that introduces AI-based technologies to speed up the discovery of new drug candidates before preclinical trials.
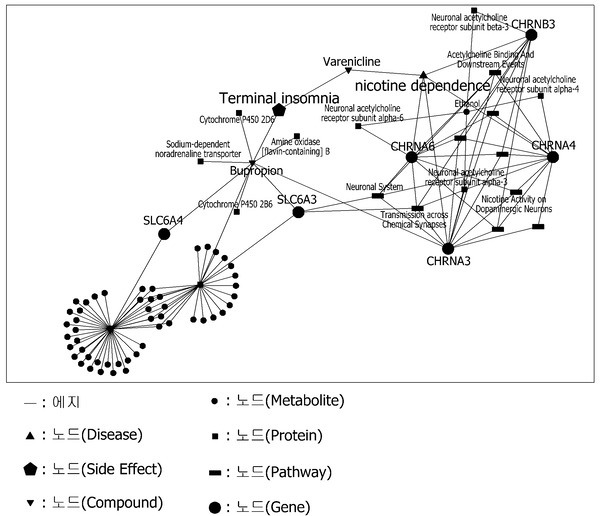
MediRita’s registered patent “Data processing apparatus and method for predicting effectiveness and safety of new drug candidate substances” (WO2020138590A1) predicts new drug candidates simply by inputting a biological network into a Convolutional Neural Network (CNN) model. Here, a biological network is a network of biological entities (e.g. genes, proteins, metabolites, symptoms, diseases, compounds and drugs) connected according to the degree of correlation between those biological entities.
Similar to what was discussed before (Atomwise’s AtomNet), the case of MediRita’s patent is also about the use of an already identified CNN model. As it is simply an application of the CNN model to predict the effect of new drugs, it lacks innovativeness. Then, what are the innovative technical features of this patent?
To examine the technical features of the invention, let’s examine the utilized data and the method for feeding the data into the CNN model.
First, I will explain the utilized data and how it is input through the overall process to predict the effect of new drug candidates.
(1) Enter a search term in the user interface unit; it can be a gene, protein, disease, compound or drug name.
Input search term.
(2) When the term is entered, biological entities related to the term and correlations between entities are extracted and a multi-omics network is created based on this. This multi-omics network is the data that is fed to the AI-based model. Here,‘omics’ is a term that refers to all technologies with the suffix ‘-omics’ that measure some characteristics of biological molecules, cells, tissues, organs, etc -such as genomics, proteomics-, while multi-omics refers to a comprehensive and integrated analysis between different levels of omics.
MediRita's Multi-omics network.
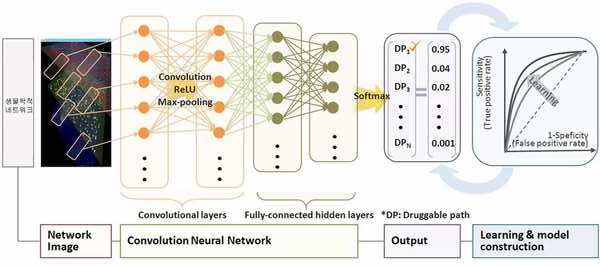
(3) By inputting the multi-omics network into the artificial neural network (ANN) model, the druggable path (DP) index for each druggable path related to the predetermined search word is obtained.
The DP index for each druggable path indicates the degree to which it is predicted to be suitable as a druggable path, selecting the paths having high DP indexes among the plurality of druggable paths.
(4) Then, how is the multi-omics network, which is the input data fed into the model?
The multi-omics network is inputted to the convolution layer as a plurality of segmented images and is sequentially input to the fully connected layer and the activation function layer to output the final result, the DP index, for each druggable path.
To conclude, MediRita’s patent predicts the effectiveness and stability of new drug candidates through CNNs, which have been proved in the field of image processing.
Although entering image data into CNNs is a technique now commonly used in many domains, I think that in the prediction of drug effects for the development of new drugs, the patentability of MediRita’s innovation was achieved through the input method of the multi-omics network into a model through processes that divide the multi-omics network into multiple pieces.
Process of obtaining DP index for each druggable path
4. Syntekabio
Syntekabio is a Korean venture company established in 2009 that uses AI-based algorithms to conduct bio business.
Syntekabio’s registered patent “Drug indication and response prediction systems and method using AI deep learning based on the convergence of different category data” (KR101953762B1) is an invention that predicts drug reactivity by inputting pharmacological functional group reactivity information and the genetic information contained in the genome into a convolution neural network (CNN) module. In other words, this patent describes a method in predicting the degree of drug response in the genome before clinical trials.
Next, through the diagram below I will explain how to train a deep learning module and how the CNN data is input to predict drug responsiveness.
A diagram illustrating an example of a merging structure of heterogeneous characteristic information for training a deep learning machine.
(1) How is the training data structured to train the model?
In the present patent, genetic information and information of drug composition become learning input data, and drug reactivity becomes labeled data (the correct answer). Therefore, the deep learning model is trained in a way that reduces the discrepancy between the output of the AI-based model and the label data during the training process.
(2) What does the input data look like?
First, the data input to the model is information on new genetic information on the mutations included in the genome and pharmacological functional information constituting the drug. The input data is in the form of a matrix as shown in the figure, wherein drugs correspond to rows and mutation features correspond to columns.
(3) What is the output data of the model?
When both the genetic information and drug composition are fed into the learning module, a result of predicting the response of the drug to the genome is obtained. In detail, the deep learning model outputs a prediction value on the drug responsiveness (IC50) of a cell line of a specific disease (such as, tumor) for the certain (input) genetic mutation information.
After reviewing Standigm’s patent, you might think that Syntekabio’s patent is similar to Standigm’s patent, since the data input into the deep learning model is also a matrix. However, Standigm patent defines genes or pathways as rows and columns as samples, whereas Syntekabio’s patent uses different types of metrics by defining information on the molecular profile of the drug as rows and genetic variation as columns. Thus, the input data of both patents are defined in different forms.
Conclusion
All of the aforementioned patents of Atomwise, Standigm, MediRita and Syntekabio, train models in a supervised learning method to predict the effects of drugs and use existing data to fit the model to obtain desired prediction values for each input data.
The effects of drugs on the human body are very complex and the amount of related biological data available to date is enormous, and as such, we could get a peek into the concern of how to obtain high accuracy predictions by inputting a large amount of data into the models and in which form.
Determining the input data is only the starting step for applying deep learning to drug discovery.
This is because to apply already known AI-based algorithms to a specific area, a new technology regarding the processing of data for the use in the specific domain must be developed.
Therefore, in the field of AI-based drug development or other applications of AI, it is expected that additional research will be conducted on the neural network structure for achieving lightweight deep learning models for the specific application, in this case, optimized for the development of new drugs.